Research Article - Modern Phytomorphology ( 2025) Volume 19, Issue 3
New biotechnological applications and metagenomic analysis of the microbial communities in seawater
Abo-Aba SEM*, Fakieh Mohammad H and Baoum Riham OAbo-Aba SEM, Biological Science Department, King Abdul-Aziz University, Jeddah, Saudi Arabia, Email: carterkelly0104@gmail.com
Received: 05-May-2025, Manuscript No. mp-25-165149; , Pre QC No. mp-25-165149 (PQ); Editor assigned: 07-May-2025, Pre QC No. mp-25-165149 (PQ); Reviewed: 21-May-2025, QC No. mp-25-165149; Revised: 07-Jul-2025, Manuscript No. mp-25-165149 (R); Published: 14-Jul-2025, DOI: 10.5281/zenodo.17062735
Abstract
Metagenomics is a method that studies genetic material extracted from environmental samples to understand the makeup, functions and dynamics of microbiomes. Studying organisms that are challenging to cultivate in laboratories and exploring species in their natural habitats are made easier by this emerging area of genetic research. These communities are diverse and widespread, found in various environments, including seawater ecosystems. Understanding their impact on ecosystems is crucial, as they include bacteria, archaea, fungi, protists and viruses. With their high challenges, seawater ecosystems have a significant microbial population, with many species playing crucial roles in biogeochemical cycles. Advancements in genomics and sequencing have revolutionized microbial ecology by using culture-independent molecular approaches. Techniques like genetic fingerprinting, metagenomics, metaproteomics, metatranscriptomics and proteogenomics provide valuable views about the microbial communities structurally and functionally, enabling studying microorganisms and their interactions with environmental elements. Overall, recognizing the implications of microbiomes on ecosystems is crucial for addressing global challenges and improving ecosystem health.
Keywords
Metagenomic, Microbiome, Biotechnological applications, Seawater microbiome
Introduction
Despite decades of research, understanding of species diversity in the diverse water-based ecosystems of our world is still limited (Alanazi et al., 2016). Microorganisms have useful genes that produce important metabolites, which are crucial for industrial and medical applications (Liem et al., 2021). They are also essential for the advancement of biotechnology and the creation of biopharmaceuticals and bioenergy (Rani et al., 2021; O’Brien 2011; Prakash 2013). Nevertheless, despite the abundance of sequencing technology tools, achieving a full knowledge of the total variety of microorganisms still poses a hard challenge. Recent research on marine biodiversity indicates that various sediments host distinct ecosystems, which are notably extreme in deep ocean habitats. Several preliminary investigations have been conducted on the utilization of marine microorganisms for the synthesis of bioactive substances, which possess diverse medical, industrialor agricultural uses (Lim et al., 2023). Metagenomics offers vital tools to investigate the variety of microbes that are "unculturable" and to search for new genes with various applications. This is possible because of the emergence of current molecular methodologies, DNA sequencing and data analysis (Singh et al., 2019).
Since 2004, extensive studies of the metagenomics of seawater have been done, revealing a remarkable range of life (Singh et al., 2020). Metagenomic investigations of the world's oceans enhance our knowledge of native microbial communities' variety, development and functional capacity (Dhakal 2017; Li et al., 2018). Metagenomics has become a crucial method in aquatic environmental research, revealing the complexities and variety of ocean microbial communities and how they interact with the environment. This methodology has greatly facilitated the identification of formerly undiscovered microorganisms, metabolic pathways and genes, significantly enhancing our comprehension of marine ecosystems and the variety of life in them (Muriel- Millán et al., 2021).
Materials and Methods
Background of biotechnology and metagenomics
In 1986, Pace and his colleagues presented an innovative concept of replicating DNA directly from environmental samples to examine the complex nature of wild microbial communities (Alves et al., 2018). Nonetheless, the word "metagenome" was introduced by a researcher named Handelsman in 1998 (Handelsman et al., 1998). Metagenomics is the scientific investigation of the genetic material present in communities of microorganisms found in the environment (Res et al., 2016). Metagenomics is sometimes referred to as ecological genomics, community genomicsand environmental genomics (Neelakanta & Sultana et al., 2013; Tseng et al., 2014). Metagenomics is a method of analyzing genomes obtained from the community of natural microorganisms without culturing (Ghosh et al., 2019). The "great plate anomaly" states that approximately 99% of the microorganisms in the surroundings cannot be cultured in the laboratory conditions. Staley and Konopka in 1985 introduced the phrase "the great plate count anomaly" to refer to the gap in the quantities of cells found in the environment that can produce surviving colonies on agar media and the quantities identified through microscopy (Harwani et al., 2012). Metagenomics offers vital tools to investigate the variety of microbes that are "unculturable" and to search for new genes with various applications. This is possible because of the emergence of current molecular methodologies, DNA sequencing and data analysis (Singh et al., 2019). This approach can read a wide range of organisms found in any environmental samples (Muriel-Millán et al., 2021).
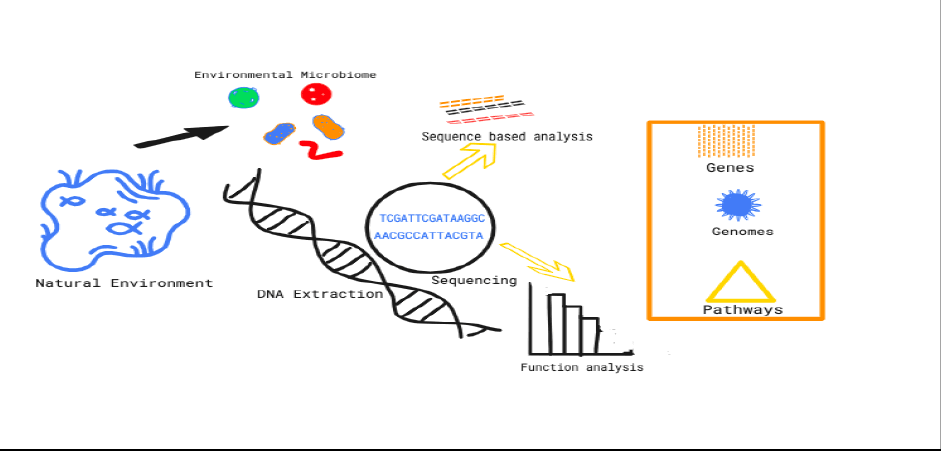
The metagenomic techniques bypasses the need for standard microbiological approaches, such as cultivation and isolation proceduresand greatly expands the range of ways microbial resources can be used (Fig. 1). It has emerged as a powerful research instrument in the fields of genetic engineering, environmental science, microbiology and biotechnology (Biddle et al., 2008). Metagenomics integrates various important advancements in molecular technology from the past century, allowing scientists to deeply investigate bacterial ecology and exploit the extensive biotechnological possibilities inside the prokaryotic population. The study of etagenomics emerged due to the realization that the diversity of prokaryotes was far larger than previously known and that the prokaryotic population held significant potential for biotechnology applications.12These results have extended beyond the limitations of traditional culture-based research (Steele et al., 2005). Several fields of study have used metagenomics, including the study of microbial populations in the human intestine, the decomposition of sugarcane bagasse waste and the examination of hypersaline environments. Furthermore, the utilization of gene resources derived from nature might provide valuable insights. Additionally, metagenomics investigations can enhance our understanding of the interconnections among microorganism populations in the biogeochemical cycle within the natural environment (Prayogo et al., 2020). The process of extracting metagenomic DNA from aquatic environments, along with functional screens, has enabled the discovery of biologically active substances, for example, enzymes and antibacterial agents, from aquatic bacteria that cannot be grown in a laboratory setting (Mahapatra et al., 2020; Park et al., 2007). In an analysis study of the functional metagenome of microbiomes above the Red Sea, they found that 59.6% of the total coding genes in the shallowest water (≤ 100 m) were bacteria, 4.16% archaea, 6.14%viruses and eukaryotes about 0.46% (Aalismail et al., 2019).
Figure 1: Diagram illustrating the metagenomic investigation study.
Results and Discussion
Metagenomics applications
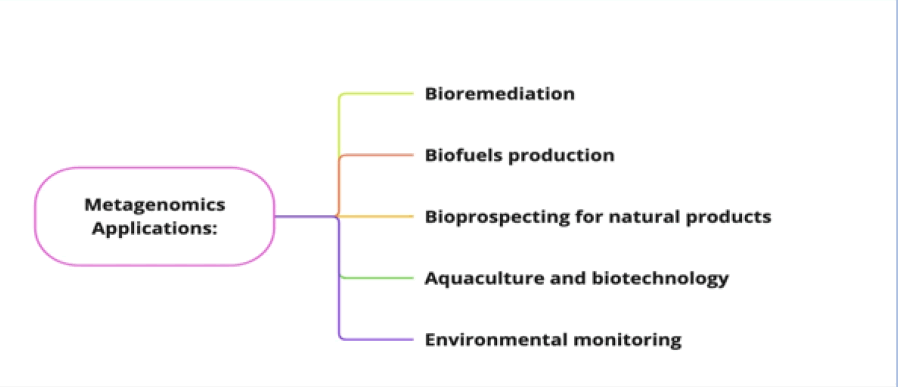
Metagenomics applications have many important and varied uses (Fig. 2). It provides innovative solutions to environmental, health and industrial challenges and enhances our comprehension of complex biological systems.
Figure 2: Applications of metagenomics. These applications support the growth of industries, the health, the economy of the society.
Bioremediation
In the environment, bioremediation is a procedure by which bacteria, fungi and plants break down, eliminate, modify, disable or detoxify various biological and toxic wastes. Microorganisms contribute to the degradation of the targeted pollutant by acting as biological catalysts and speeding up chemical reactions using their enzymatic pathways. Microorganisms can only combat contaminants if they have access to a wide range of chemicals that can provide them with energy and nutrients for cell growth.
Aquatic environments are highly challenging conditions on Earth and harbor a large proportion of the worldwide bacterial groups. Certain bacterial species in these environments have crucial functions in several biogeochemical pathways (Muriel-Millán et al., 2021). Oceans and seas cover more than 70% of the Earth's surface area. These large bodies of water play an important role in regulating the global weather and are responsible for producing 50% of the surrounding oxygen (Bollmann et al., 2010). Bacteria and archaea occupy almost all aquatic ecosystems, with an estimated abundance of 5.5 × 1029 cells in oceans, that includes both water and surface and deep-sea sediments, constituting up to 90% of total biomass from the oceans (Flemming et al., 2019). Marine sediments host a significant fraction of Earth's bacterial biomass, presented to have a worldwide bacterial diversity of 32,800 species (Hoshino et al., 2020). Factors including oxygen levels, distance from surface sediment and carbon levels affect the bacterial makeup of the ocean bottom (Hoshino et al., 2020). Multiple studies have shown that a community of bacteria might be more effective in bioremediation compared to individual species due to their inability to break down all the chemicals present in crude oil (Santisi et al., 2019). Since of the toxicity of raw oil, oil spillage have a significant impact on the ecological system. While specific chemical and physical procedures are utilized to clean up oil-contaminated locations, biologically based methods are more environmentally sustainable and productive (Mapelli et al., 2017).Bacteria species in marine environments from the Alcanivorax, Cycloclasticus and Marinobacter genera have been isolated from various environments and are well known to be effective hydrocarbon degraders. Other marine species are now being discovered to decompose various chemicals efficiently, demonstrating specific characteristics like breakdown a wide variety of pH readings, temperatures and NaCl levels, as well as bio-surfactant synthesis (Muriel-Millán et al., 2021). Biological processes are unable to eliminate heavy metals ("no degradation"), as they just change their nuclear structure. However, these metals can be converted from one state of oxidation or organic complex to a different one. Microorganisms have evolved numerous processes, including adsorption, absorption, methylation, oxidation and reduction, to protect themselves from the harmful effects of heavy metal toxicity. Microorganisms actively accumulate toxic substances through metabolism or passively through adsorption. Microbial methylation is crucial in the bioremediation of harmful substances due to the frequent volatility of methylated molecules (Abatenh et al., 2017).
Biofuels production
Recently, the interest in producing biofuel using microbes has constantly grown. To deal with the insufficient worldwide energy supplies and address worries about global warming, significant endeavors have been undertaken to minimize the world's reliance on traditional petroleum fuel reserves and develop cutting-edge technology to produce sustainable fuels (Patel et al., 2015). Microbial-derived biofuels, including biodiesel, ethanol and butanol, have become an appealing replacement and received increased attention from scientists, industry and governments because of their environmental benefits (Xing et al., 2012). At present, about 99.9% of microorganisms are incapable of detection or isolation from their natural habitats, which poses a significant constraint on the traditional approach to enzyme manufacturing that relies on cultivating bacterial isolates. Therefore, extensive research is currently being conducted globally to utilize unexplored microbial ecosystems to identify and isolate powerful enzymes (Liu et al., 2022). In the field of biofuels, metagenomics is playing a big role in identifying new enzymes that break down cell walls. These enzymes are crucial for breaking down lignocellulosic biomass, which can then be converted into biofuel. This discovery is a cost-effective method for advancing biofuel technology (Sartaj et al., 2022). The following enzymes have been identified via a metagenomic approach with their industrial applications. Xylanase from sea bottom as bioethanol (Hung et al., 2011), Esterase from antarctic soil as bioethanol production (Fu et al., 2011), and Laccase from water from the South China Sea as a dye degradation (Fang et al., 2011). A remarkable lipase enzyme identified as LipEH166 has been discovered in metagenomic libraries extracted from intertidal flat sediments in South Korean coastal locations. This enzyme plays an important role in the synthesizing of biodiesel and is also utilized for the bioremediation of lipid-rich wastewater collected from various industries, including dairy, cosmetics, food and detergents (Kim et al., 2009).
Bioprospecting for natural products
Bioprospecting indicates the scientific research for innovative biological and genetic resources found in plants, animals and microbes using biotechnological applications in their natural environments (Lozada et al., 2015). The fundamental concept is that conducting such a search has the potential to reveal genes and chemical compounds that can greatly help humanity, particularly through delivering medications (Ten Have et al., 2021). A variety of enzymatic activities has been detected in cultivated marine microbes, indicating the possibility of uncovering new enzymes from marine bacteria that are distributed across many regions of the world (Madhavan et al., 2017).
Many researchers isolated microorganisms producing enzymes such as protease, the biggest group of enzymes which are commercially useful in bio-industries such as tanneries, the industry of food, washing powders, processing of leather, medicines, molecular biological studies and peptide synthesis (Kumar et al., 1999), keratanase which are particular proteolytic enzymes that can degrade insoluble keratins (Brandelli et al., 2010), and amylase an important enzyme for use in the industrial starch conversion process (Nigam et al., 1995). The technique of bioprospecting marine bacteria involves an organized search for valuable and significant products that can benefit society, such as genes, proteins and secondary metabolites (López et al., 2019). Microorganisms offer a varied range of natural compounds that can be used to develop novel medications for treating human diseases such as infections and cancer. More than 50% of pharmaceutical medications on the market were developed or driven by natural products (Newman et al., 2016).
Aquaculture and biotechnology
Aquaculture is a technique used to provide food and other goods, restore ecosystems and replenish wild populations, as well as revive species that are at risk of extinction. Aquaculture (mariculture) is now recognized as the most rapidly developing area of the global agricultural industry. Due to environmental degradation, there has been a critical transition in marine farming production from the coast to the deep regions due to the presence of illnesses and the high density of mariculture (Li et al., 2023). To supply edible marine products to the growing global need for protein, aquaculture is becoming increasingly important (Cui et al., 2022). However, the rapid increase in aquaculture production gives rise to several safety problems, including both environmental and human health aspects. Considering the significant societal and financial consequences of this activity, it is crucial to recognize the ecological and healthcare concerns linked to it (Haro-Moreno et al., 2020). Nevertheless, the widespread application of chemicals and the buildup of organic material, such as excessive feed and fish waste, result in the pollution of the water layers, increase and modifying the physicochemical parameters of the water (Martinez-Porchas et al., 2012). These changes in water characteristics can severely impact the environment or modifying the interrelated marine microbial ecosystems (Martinez-Porchas et al., 2017).
Environmental monitoring
Ecological stress and human behaviors, such as air pollution, have effectively limited or stimulated the development of< specific microbial communities (Fig. 3). According to the information provided, numerous research has described the use of microbes as biosensors to detect changes in the ecosystem (Gavrilas et al., 2022). In the past few years, biosensors have undergone rapid and varied development (Nigam et al., 2015). Microbial biosensors are devices for analysis that detects compounds in the environment through specific biological responses of the microbe or its components (Vogrinc et al., 2015).Environmental microbiology has advanced to efficiently collect vital bio-monitoring data through independent molecular culture technologies. Molecular approaches such as quantitative Polymerase Chain Reaction (PCR), high-throughput sequencing, PhyloChip, GeoChip and metagenomics have considerably increased our understanding of the variety and functional< heterogeneity of marine microbial populations (Sharma et al., 2022).
Figure 3: Metagenomic analysis of microbial communities underwater for environmental monitoring applications.
Conclusion
Metagenomics is a new way to study the genome of the microbial world. It is the scientific investigation of the genetic material and metabolic variations derived from communities of microorganisms present in the environment. Studying the metagenome of marine ecosystems can uncover unique sets of genes that are associated with specific habitats, such as the deep water and surface. Metagenomics effectiveness has been demonstrated in enzymes and metabolic pathways, significantly enhancing our knowledge of< microbial ecology in the natural environment. The presented review paper was conducted to facilitate an in-depth understanding of metagenomics. Metagenomics has been utilized to assist in discovering new enzymes in nature that may be utilized for bioremediation, biofuels like biodiesel and other industrial applications.
Metagenomics has become a highly intriguing approach for investigating the range of microorganisms in aquatic habitats that can produce antibiotics which can help reduce the contamination in aquaculture.
Author Contributions
Conceptualization, Abo-Aba Salah El-Deen M; Abo-Aba M, Fakieh Mohammad H, Baoum Riham O, writing review and editing; All writers have read and approved the final version of the manuscript.
Data Availability
All the data used in this study is included in the paper. All diagrams were created by www.autodraw.com/; www.firefly.adobe.com; and https://miro.com.
Funding
No funding.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
None.
Acknowledgments
None.
CS Statement
This article does not include any studies involving humans or animals done by the writers.
References
- Liem M, Regensburg-Tuïnk T, Henkel C, Jansen H, Spaink H. (2021). Microbial diversity characterization of seawater in a pilot study using Oxford Nanopore Technologies long-read sequencing. BMC Res Notes. 14:1-7.
[Crossref] [Google Scholar] [PubMed]
- Rani A, Saini KC, Bast F. (2021). Microorganisms: A potential source of bioactive molecules for antioxidant applications. Molecules. 26:1142.
[Crossref] [Google Scholar] [PubMed]
- O’Brien J, Wright GD. (2011). An ecological perspective of microbial secondary metabolism. Curr Opin Biotechnol. 22:552-558.
[Crossref] [Google Scholar] [PubMed]
- Prakash O, Shouche Y, Jangid K, Kostka JE. (2013). Microbial cultivation and the role of microbial resource centers in the omics era. Appl Microbiol Biotechnol. 97:51-62.
[Crossref] [Google Scholar] [PubMed]
- Lim Y, Yang SJ, Kang I, Cho JC. (2023). Metagenomic data from surface seawater of the east coast of South Korea. Scientific Data. 10:1-7.
[Crossref] [Google Scholar] [PubMed]
- Singh BP, Rateb ME, Rodriguez-Couto S, Polizeli MD, Li WJ. (2019). Editorial: Microbial secondary metabolites: Recent developments and technological challenges. Front Microbiol. 10:454593.
[Crossref]
- Singh S, Singh H, Rout B, Tripathi RBM, Chopra C, Chopra RS. (2020). The new science of metagenomics: Revealing the secrets of microbial physiology. Metagenomics: Techniques, Applications, Challenges and Opportunities. 3-22.
- Dhakal D, Pokhrel AR, Shrestha B, Sohng JK. (2017). Marine rare actinobacteria: Isolation, characterization, and strategies for harnessing bioactive compounds. Front Microbiol. 8:251578.
[Crossref] [Google Scholar] [PubMed]
- Sunagawa S, Coelho LP, Chaffron S. (1979). Structure and function of the global ocean microbiome. Science. 348.
[Crossref] [Google Scholar] [PubMed]
- Li Y, Jing H, Xia X, Cheung S, Suzuki K, Liu H. (2018). Metagenomic insights into the microbial community and nutrient cycling in the western subarctic Pacific Ocean. Front Microbiol. 9:337314.
[Crossref] [Google Scholar] [PubMed]
- Muriel-Millán LF, Millán-López S, Pardo-López L. (2021). Biotechnological applications of marine bacteria in bioremediation of environments polluted with hydrocarbons and plastics. Appl Microbiol Biotechnol. 105:7171-7185.
[Crossref] [Google Scholar] [PubMed]
- Alves LDF, Westmann CA, Lovate GL, de Siqueira GMV, Borelli TC, Guazzaroni ME. (2018). Metagenomic approaches for understanding new concepts in microbial science. Int J Genomics.
[Crossref] [Google Scholar] [PubMed]
- Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. (1998). Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem Biol. 5.
[Crossref] [Google Scholar] [PubMed]
- Nazir A. (2016). Review on metagenomics and its applications. Imp J Intersd Res. 2.
- Neelakanta G, Sultana H. (2013). The use of metagenomic approaches to analyze changes in microbial communities. Microbiol Insights. 6:37.
[Crossref] [Google Scholar] [PubMed]
- Tseng CH, Tang SL. (2014). Marine microbial metagenomics: From individual to the environment. Int J Mol Sci. 15:8878-8892.
[Crossref] [Google Scholar] [PubMed]
- Ghosh A, Mehta A, Khan AM. (2019) Metagenomic analysis and its applications. Encyclopedia of Bioinformatics and Computational Biology: ABC of Bioinformatics. 1-3:184-193.
- Harwani D. (2012). The great plate count anomaly and the unculturable bacteria. Int J Sci Res. 2:350-351.
- Biddle JF, Fitz-Gibbon S, Schuster SC, Brenchley JE, House CH. (2008). Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc Natl Acad Sci U S A. 105:10583-10588.
[Crossref] [Google Scholar] [PubMed]
- Steele HL, Streit WR. (2005). Metagenomics: Advances in ecology and biotechnology. FEMS Microbiol Lett. 247:105-111.
[Crossref] [Google Scholar] [PubMed]
- Prayogo FA, Budiharjo A, Kusumaningrum HP, Wijanarka W, Suprihadi A, Nurhayati N. (2020). Metagenomic applications in exploration and development of novel enzymes from nature: A review. J Genet Eng Biotechnol. 18:1-10.
[Crossref] [Google Scholar] [PubMed]
- Mahapatra GP, Raman S, Nayak S, Gouda S, Das G, Patra JK. (2020). Metagenomics approaches in discovery and development of new bioactive compounds from marine actinomycetes. Curr Microbiol. 77:645-656.
[Crossref] [Google Scholar] [PubMed]
- Park HJ, Jeon JH, Kang SG, Lee JH, Lee SA, Kim HK. (2007). Functional expression and refolding of new alkaline esterase, EM2L8 from deep-sea sediment metagenome. Protein Expr Purif. 52:340-347.
[Crossref] [Google Scholar] [PubMed]
- Aalismail NA, Ngugi DK, Díaz-Rúa R, Alam I, Cusack M, Duarte CM. (2019). Functional metagenomic analysis of dust-associated microbiomes above the Red Sea. Sci Rep. 9:1-12.
[Crossref] [Google Scholar] [PubMed]
- Bollmann A, Palumbo AV., Lewis K, Epstein SS. (2010). Isolation and physiology of bacteria from contaminated subsurface sediments. Appl Environ Microbiol. 76:7413-7419.
[Crossref] [Google Scholar] [PubMed]
- Flemming HC, Wuertz S. (2019). Bacteria and archaea on Earth and their abundance in biofilms. Nat Rev Microbiol. 17:247-260.
[Crossref] [Google Scholar] [PubMed]
- Hoshino T, Doi H, Uramoto GI. (2020). Global diversity of microbial communities in marine sediment. Proc Natl Acad Sci U S A. 117:27587-27597.
[Crossref] [Google Scholar] [PubMed]
- Santisi S, Catalfamo M, Bonsignore M. (2019). Biodegradation ability of two selected microbial autochthonous consortia from a chronically polluted marine coastal area (Priolo Gargallo, Italy). J Appl Microbiol. 127:618-629.
[Crossref] [Google Scholar] [PubMed]
- Mapelli F, Scoma A, Michoud G. (2017). Biotechnologies for marine oil spill cleanup: Indissoluble ties with microorganisms. Trends Biotechnol. 35:860-870.
[Crossref] [Google Scholar] [PubMed]
- Abatenh E, Gizaw B, Tsegaye Z, Wassie M. (2017). The role of microorganisms in bioremediation- a review. Open J Environ Biol. 2:038-046.
[Crossref]
- Patel A, Sarkar O, Rova U, Christakopoulos P, Matsakas L. (2021). Valorization of volatile fatty acids derived from low-cost organic waste for lipogenesis in oleaginous microorganisms-A review. Bioresour Technol. 321:124457.
[Crossref] [Google Scholar] [PubMed]
- Xing MN, Zhang XZ, Huang H. (2012). Application of metagenomic techniques in mining enzymes from microbial communities for biofuel synthesis. Biotechnol Adv. 30:920-929.
[Crossref] [Google Scholar] [PubMed]
- Liu S, Moon CD, Zheng N, Huws S, Zhao S, Wang J. (2022). Opportunities and challenges of using metagenomic data to bring uncultured microbes into cultivation. Microbiome. 10:1-14.
[Crossref] [Google Scholar] [PubMed]
- Sartaj K, Patel A, Matsakas L, Prasad R. (2022). Unravelling metagenomics approach for microbial biofuel production. Genes. 13:1942.
[Crossref] [Google Scholar] [PubMed]
- Hung KS, Liu SM, Tzou WS. (2011). Characterization of a novel GH10 thermostable, halophilic xylanase from the marine bacterium Thermoanaerobacterium saccharolyticum NTOU1. Process Biochem. 46:1257-1263.
- Fu C, Hu Y, Xie F. (2011). Molecular cloning and characterization of a new cold-active esterase from a deep-sea metagenomic library. Appl Microb Biotechnol. 90:961-970.
[Crossref] [Google Scholar] [PubMed]
- Fang Z, Li T, Wang Q. (2011). A bacterial laccase from marine microbial metagenome exhibiting chloride tolerance and dye decolorization ability. Appl Microbiol Biotechnol. 89:1103-1110.
[Crossref] [Google Scholar] [PubMed]
- Kim EY, Oh KH, Lee MH, Kang CH, Oh TK, Yoon JH. (2009). Novel cold-adapted alkaline lipase from an intertidal flat metagenome and proposal for a new family of bacterial lipases. Appl Environ Microbiol. 75:257.
[Crossref] [Google Scholar] [PubMed]
- Lozada M, Dionisi HM. (2015). Microbial bioprospecting in marine environments. Springer Handbook of Marine Biotechnology. 307-326.
- ten Have H, Patrão Neves MD. (2021). Bioprospecting (See Biopiracy). Dictionary of Global Bioethics. 197-198.
- Madhavan A, Sindhu R, Parameswaran B, Sukumaran RK, Pandey A. (2017). Metagenome analysis: A powerful tool for enzyme bioprospecting. Appl Biochem Biotechnol. 183:636-651.
[Crossref] [Google Scholar] [PubMed]
- Kumar CG, Takagi H. (1999). Microbial alkaline proteases: from a bioindustrial viewpoint. Biotechnol Adv. 17:561-594.
[Crossref] [Google Scholar] [PubMed]
- Brandelli A, Daroit DJ, Riffel A. (2010). Biochemical features of microbial keratinases and their production and applications. Appl Microbiol Biotechnol. 85:1735-1750.
[Crossref] [Google Scholar] [PubMed]
- Nigam P, Singh D. (1995). Enzyme and microbial systems involved in starch processing. Enzyme Microb Technol. 17:770-778.
- López LP. (2019). Marine bioprospecting. Marine and fisheries policies in Latin America: A comparison of selected countries. 33-44.
- Newman DJ, Cragg GM. (2016). Natural products as sources of new drugs from 1981 to 2014. J Nat Prod.79:629-661.
[Crossref] [Google Scholar] [PubMed]
- Li S, Wang S, Pan C. (2023). Differences in physiological performance and gut microbiota between deep-sea and coastal aquaculture of Thachinotus ovatus: A metagenomic approach. Animals (Basel). 13:3365.
[Crossref] [Google Scholar] [PubMed]
- Cui G, Liu Z, Xu W. (2022). Metagenomic exploration of antibiotic resistance genes and their hosts in aquaculture waters of the semi-closed Dongshan Bay (China). Sci Total Environ. 838:155784.
[Crossref] [Google Scholar] [PubMed]
- Haro-Moreno JM, Coutinho FH, Zaragoza-Solas A, Picazo A, Almagro-Moreno S, López-Pérez M. (2020). Dysbiosis in marine aquaculture revealed through microbiome analysis: reverse ecology for environmental sustainability. FEMS Microbiol Ecol. 96.
[Crossref] [Google Scholar] [PubMed]
- Martinez-Porchas M, Martinez-Cordova LR. (2012). World aquaculture: Environmental impacts and troubleshooting alternatives. Sci World J. 2012.
[Crossref] [Google Scholar] [PubMed]
- Martínez-Porchas M, Vargas-Albores F. (2017). Microbial metagenomics in aquaculture: A potential tool for a deeper insight into the activity. Rev Aquac. 9:42-56.
- GavrilaÈ S, Ursachi C Ètefan, PerÈa-CriÈan S, Munteanu FD. (2022). Recent trends in biosensors for environmental quality monitoring. Sensors (Basel). 22.
[Crossref] [Google Scholar] [PubMed]
- Nigam VK, Shukla P. (2015). Enzyme based biosensors for detection of environmental pollutants-a review. J Microbiol Biotechnol. 25:1773-1781.
[Crossref] [Google Scholar] [PubMed]
- Vogrinc D, Vodovnik M, Marinšek-Logar R. (2015). Microbial biosensors for environmental monitoring. Acta Agric Slov. 106:67-75.
- Sharma P, Bano A, Singh SP, Dubey NK, Chandra R, Iqbal HMN. (2022). Microbial fingerprinting techniques and their role in the remediation of environmental pollution. Cleaner Chem Eng. 2:100026.