Research Article - Modern Phytomorphology ( 2025) Volume 19, Issue 4
Genetic Transformation of Tomato with Enhanced Resistance against Fusarium Wilt Using Mulberry Chitinase Gene
Mubarak Ali Anjum1*, Muhammad Ashfaq2*, Usman Arif3, Muhammad Tariq3, Farah Naz3, Rabiah Ashraf3, Abdul Munim Farooq3, Muhammad Ali4, Saleem Haider1, Muhammad Atif5 and Qurban Ali22Department of Plant Breeding and Genetics, Faculty of Agricultural Sciences, University of the Punjab, Lahore, Pakistan
3Center of Excellence in Molecular Biology, University of the Punjab, Lahore, Pakistan
4Department of Entomology, Faculty of Agricultural Sciences, University of the Punjab, Lahore, Pakistan
5Center of Applied Molecular Biology, University of the Punjab, Lahore, Pakistan
Mubarak Ali Anjum, Department of Plant Pathology, Faculty of Agricultural Sciences, University of the Punjab, Lahore, Pakistan, Email: mubarak.ali.anjum1103@gmail.com Muhammad Ashfaq, Department of Plant Breeding and Genetics, Faculty of Agricultural Sciences, University of the Punjab, Lahore, Pakistan, Email: ashfaq.iags@pu.edu.pk
Received: 18-Jun-2025, Manuscript No. mp-25-166963; , Pre QC No. mp-25-166963 (PQ); Editor assigned: 20-Jun-2025, Pre QC No. mp-25-166963 (PQ); Reviewed: 04-Jul-2025, QC No. mp-25-166963; Revised: 11-Jul-2025, Manuscript No. mp-25-166963 (R); Published: 18-Jul-2025, DOI: 10.5281/zenodo.17129941
Abstract
Different strategies have been adopted to control diseases in tomato crops caused by various fungal pathogens. These are cultural, biological, chemical and biotechnological approach. Recently biotechnological approach is gaining attention as suitable control measures for disease resistance. Gene transformation in the plant to improve resistance is effective measures. Chitinase genes effectively increase resistance in plants against fungal pathogens and insects. In this study a chitinase gene MnChi10 from mulberry (Morus notabilis) tree was isolated, characterized, amplified and transformed into tomato plant for effective control of Fusarium wilt disease. Chitinase MnChi10 gene showed homology with family 19 of glycosyl hydrolyses. Furthermore, the MnChi10 gene was transformed and expressed in tomato plants through a modified Agrobacterium-mediated transformation. Transformed tomato plants showed higher chitinase activity than control plants, as well as better growth and increased resistance against Fusarium wilt disease. It was observed that transformed plants suppressed fungal growth and continued to thrive even under heavy fungal infestation. It is concluded that chitinase gene MnChi10 showed promising results in resistance against Fusarium wilts. These tomato plants, in future, may be used as resistant variety for commercial growing.
Keywords
Tomato, Transgenic resistance, Chitinase, MnChi10, pCAMBIA 1301, Fluorescent In Situ Hybridization
Introduction
Crop plants are susceptible to biotic and abiotic factors which are pathogen, diseases and stress. These factors are involved in yield reduction of crops and affect quality and quantity of agricultural commodities (Mwangi, et al. 2023). Crop diseases are most economical and devastating pathogens (Doehlemann, et al. 2017). Fungi are notorious and most dominant casual agents of plant diseases. Among these notorious fungi, some can kill their host while others are not (Fauteux, et al. 2005). Crops mainly affected by fungi are tomato, potato, cucumber, pepper, cotton, wheat and other crops (Thambugala, et al. 2020, Panno, et al. 2021). Tomato (Solanum lycopersicum L.) is an important and extensively grown crop of Mediterranean region throughout the world (Brahimi, et al. 2017). Tomatoes are a good source of important nutrients and minerals. These are lycopene, potassium, iron, folate and vitamin C (Staniaszek, et al. 2014). Besides vitamins, tomatoes are also a source of antioxidants. Β-carotene, phenolic compounds (flavonoids), hydroxycinnamic acid, homovanillic acid, ferulic acid and chlorogenic (Ullah, et al. 2017). Tomato crop is grown all over the world as winter and summer vegetable and is prone to various serious diseases i.e. attacked by a series of pathogens (Collins, et al. 2022). Tomato crop is highly susceptible to attack by fungal pathogens, Fusarium and Alternaria (Nazarov, et al. 2020). Fusarium oxysporum is an important pathogen, soil fungus and exists as saprophyte. It has the ability to degrade lignin and is a endophyte of plant roots (Chitwood-Brown, et al. 2021). More than 120 different forms of Fusarium oxysporum are commonly found causing various diseases (Arie, 2019). Fusarium oxysporum penetrates into plant roots and colonize the vascular tissues (Wang, et al. 2020). There are no effective chemical control measures for Fusarium wilts (Díaz and Jiménez-Gasco, 2011). In previous era, when the agriculture is started, the growers had to tackle the damaging organisms. These damaging organisms are controlled by physical and biological ways (Recep, et al. 2009). In developing countries, chemical control pollutes the water with poisons and carcinogenic materials. Multipurpose pesticides are available in the market that not only destroys target pathogen but also beneficial species (Chaerani and Voorrips, 2006).
To control devastating effects of wilts fungi, biotechnological approach is mandatory through genetic modifications (Anderson, et al. 2004). The second most abundant biopolymer in the earth is chitin, composed of β 1, 4-N-acetylglucosamine (GlcNAC) (Hamid, et al. 2013). Outer skeleton of insects, crabs, lobsters, shrimps and cell wall of fungi, yeast, algae while the internal structure of few invertebrates is made of chitin (Rathore and Gupta, 2015). Chitin occurs in three polymorphic forms i.e. Alpha chains are arranged in antiparallel fashion. Beta chains are arranged in parallel fashion while in the gamma form of chitin chains are arranged in both forms. Alpha chains are most abundant forms of chitin (Wubie, et al. 2014). Chitin is degraded by chitinases in two steps, first the chitin oligosaccharides are formed by cleavage of chitinase while in second step further cleavage cause formation of N-acetylglucosamine and monosaccharide (García, et al. 2001). About 16% of dry weight of filamentous fungi and basadiomycetes consists of chitin. The precursor for synthesis of chitin is uridine diphosphate N-acetyl-D-glucosamine that synthesize chitin by the help of enzyme chitin synthase (Suginta, et al. 2000). As chitin is important part of the cell wall of fungi there must be a system by which it can be break down and remolded to give it some degree of plasticity that is important in budding and elongation of hyphae (Dana, et al. 2006). Chitinolytic enzymes are categorized in 3 families of glycosyl hydrolases i.e., family 18, 19 and 20. This grouping is done on sequence similarity of amino acids (Kumar, et al. 2018).
The plant chitinases are endo-chitinases and are present in stem, seed, tuber and flowers. Plant chitinases are categorized in 5-6 classes based on amino acid structure (Cletus, et al. 2013). Plant chitinases are produced in response to the pathogen or contacting with elicitor or growth regulators. There are reports that chitinases are expressed in response to abiotic stress such as high salt concentration, drought and cold (Sivaji, et al. 2014). There are different roles of cellular and secretory chitinases in plant defense pathway. Apo plastic chitinases block the growth of fungal hyphae by induced response and are considered as part of initial response (Cao J and Tan, 2019). It also stimulates the defense pathway by releasing of fungal elicitors. Chitinases are released when the fungal hyphae invade, penetrate and affect cell integrity (Collinge, et al. 2008). Class I chitinase are vacuolar and are induced by ethylene and jasmonate pathway while chitinase II are induced systematic response (Davis, et al. 2002). The role of chitinases in defense against fungi has been strongly established by genetic transformation experiments (Di, et al. 2016). The role of chitinase gene transformation in tomato plant is documented to control fungal pathogens. Resistance in tomato plant is built up by genetic engineering techniques against fungal pathogens but there is a lack of data related to chitinase genes confers resistance against fungal pathogens (Gai, et al. 2017). The main objectives of our study were to isolate and characterize the chitinase gene from mulberry tree (Morus notabilis), development of transgenic tomato plants and analyze the overall resistance of transgenic tomato plants against wilt fungi. In future, the project will help to understand the use of chitinase gene resistance against fungal pathogens.
Materials and Methods
Sample collection
Mulberry (Morus notabilis) tree leaves based on morphology are collected from University of the Punjab, Canal Road Lahore, Pakistan.
Extraction and purification of Mulberry DNA
Total DNA from mulberry leaves is extracted by CTAB (Cetyltrimethyl ammonium bromide) method and purified.
Amplification of chitinase gene
Chitinase gene extracted from mulberry tree leaves was amplified by using a specific set of primers listed below (Tab. 1).
| Primers | Sequence | Product size |
|---|---|---|
| Pair 01 | CGCCAACACCTTCTACACCT | 982 BP |
Table 1. List of Primer for mulberry chitinase gene
Cloning of citinase gene in pET30a vector
For mulberry chitinase gene cloning in prokaryotic expression vector, the chitinase gene primers was indulged with Nco1 and BgIII sites in both reverse and forward. pET30a vector cloning is used for protein isolation and characterization.
Cloning of chitinase gene is pCAMBIA 1301 vector
Amplification of chitinase primer for expression: The mulberry chitinase gene was optimized and synthesized for transformation in tomato variety Rio Grande. The codon optimized gene and promoter 35 s upstream and the GUS reporter downstream was synthesized from BioBasic Canada in multiple cloning sites of pCAMBIA 1301 with HindIII and EcoR1 enzymes were used. Transformation of chitinase gene construct in Agrobacterium: Agrobacterium strain LB4404 was used for chitinase gene transformation in tomato plant.
Agrobacterium competent cells preparation
Agrobacterium strain LB4404 was used in competent cells making and transforming in pCAMBIA 1301 plasmid with chitinase gene in it. YEP media is prepared and single colony was inoculated on it, placed it for 48 hours. YEP broth was diluted and incubated for 600 nm OD at 30°C. Centrifuge the cells for 15 minutes, the pellet was washed with HEPES 1 mM. It was washed with chilled 10% glycerol for 10 minutes. After washing, the pellet was suspended in YEP broth containing glycerol with final concentration of 10%.
Electroporation of chitinase gene construct in Agrobacterium
Bio-Rad Electroporation device (#165-2105) was used for Electroporation of chitinase gene containing plasmid in Agrobacterium. Electroporation device was set to capacitance of 25 μF, 2.2 KV voltage and resistance of 200 ohms for a constant time. 80 μl of competent cells were mixed with 100 ng or 3 μl construct and place it on ice. 500 μl of YEP broth was immediately added to cuvette and placed on ice for 5 minutes. Thereafter the cuvette was shifted in a fresh vial and incubated for 2 hours at 30°C. YEP agar containing kanamycine is prepared and transformed culture was spread on it and kept for 48 hours at 30°C.
Transformations of tomato plant with chitinase construct
After confirmation of vector transformation, a single node of tomato cultivar Rio-Grande was used in transformation of construct.
Dissecting of intermodal part of tomato plant stem
A node of tomato plant was dissected with sterile surgical blade in a laminar flow chamber. About 1 cm of part was prepared as explants for transformation.
Agrobacterium mediated transformation
The Agrobacterium strain LB4404 containing chitinase gene plasmid was streaked on solidified agar medium containing 50 μg/ml kanamycin and incubated at 30°C for 24 to 48 hours. A single colony was inoculated in 5 ml YEP broth containing kanamycin in 50 μg/ml concentration in 50 ml culture tube. The samples were incubated at 30°C for 24 hours with 200 rpm on rotary shaker. After incubation, the bacterial suspension was directly used in co-cultivation with internodes of the stem. The tubes were placed on the orbital shaker for 30 minutes. The internodes were dried on blotting paper, placed on MS plaates and kept in dark for 48 hours. After dark 48 hours, the internodes were transferred to MS plates containing cefatoxime 250 μg/ml antibiotics and allowed to grow for 6-8 weeks.
Molecular analysis of transformed tomato cultivar
Regenerated tomato plantlets were multiplied by single node cuttings onto MS basal medium and subsequently analysed for sorting positive transgenic plants and PCR.
Antifungal analysis of transgenic plantlets
In-vitro fungal bioassay: The experiments were further analyzed for antifungal activity against already grown Fusarium oxysporum on potato dextrose agar. A block of agar 5 mm with mycelium growth was placed on base of 4 week old transgenic plants and non-transgenic plants. The inoculated vessels were incubated at room temperature for 3 weeks. The experiment was repeated in a triplicate expression of chitinase gene.
Real time PCR
The best grown plants were used for expression at different intervals at 0 h, 12 h, 24 h, 36 h, 48 h and 72 hours when inoculated with Fusarium oxysporum. RNA was isolated and cDNA was synthesized. SYBR green dye (ROX, qPCR Master Mix 2X) was used. The threshold cycle (Cq) was normalized to the Cq of housekeeping gene, namely beta actin and was amplified with the samples.
Fluorescent in situ hybridization (FISH) analysis
Probe labeling and detection: Probe for fluorescence is labeled for chitinase gene detection.
Results
The study aims to develop transgenic tomato lines tolerant to wilt fungi (Fusarium oxysporum). Codon optimized chitinase gene was used in the study to evaluate gene expression.
Detection and identification of mulberry chitinase gene
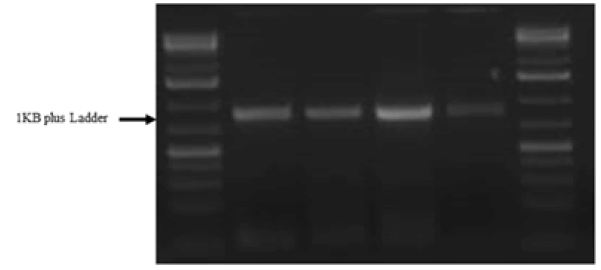
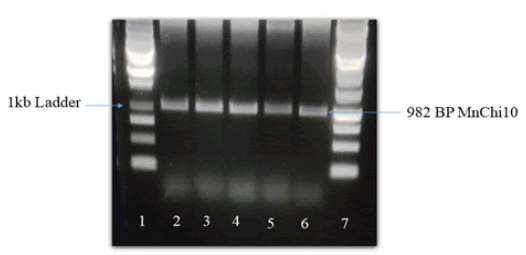
Chitinase gene of 982 BP was isolated and characterized (Fig. 1).
Figure 1. Amplified chitinase gene of mulberry RNA, Band size of 982 BP. Lane 1=1 kb plus ladder, Lane 2-4=samples.
Recombinant chitinase gene expression
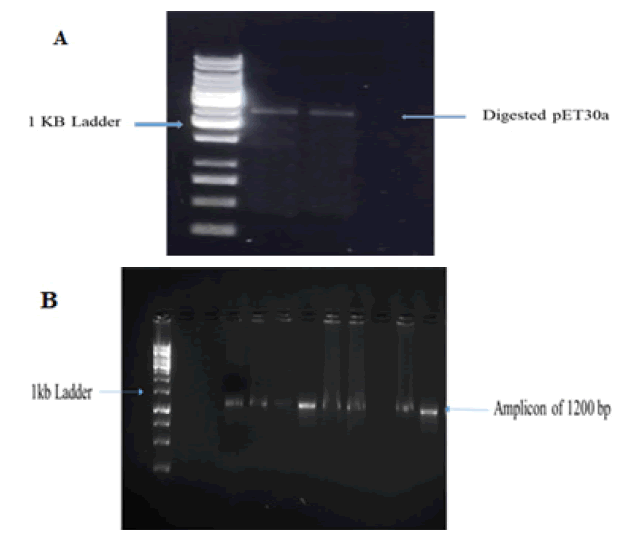
Full length 982 BP mulberry chitinase gene was successfully amplified. Sequence of chitinase gene from recombinant TA clone was analyzed and submitted in gene Bank NCBI with accession number PP951523. Fig. 2 indicates restriction digestion of mulberry chitinase gene in pET30a vector.
The amplified mulberry chitinase gene was cloned directionally in pET30a vector with T7 promoter and restriction enzymes BgIII and NcoI were used. Successful cloning was indicated by digestion of recombinant plasmid of pET30a.
Figure 2: (A) Restriction digestion by EcoR1 and BamH1, Lane 1=1 kb ladder, Lane 2-3=samples of digested vector, (B) Confirmation of chitinase gene ligation in pET30a vector.
Mulberry chitinase gene transformation in tomato plant
Rio-grande tomato variety was transformed with mulberry chitinase gene, MnChi10 via Agrobacterium mediated transformation. Fig. 3 illustrated regeneration, transformation and development of tomato plants. A total of 400 plant nodes were transformed by Agrobacterium mediated transformation. Agrobacterium strain used was LB4404. A total of 180 plantlets regenerated into complete plants. The transformed regenerated plants were confirmed by PCR amplification of chitinase gene in PCR reaction.
Figure 3: Steps in tomato transformation and regeneration. (A) Tomato internodes (B) Regeneration of tomato internodes after transformation in MS medium (C) Acclimatized tomato plants in pots.
Fish and molecular analysis of transformed tomato plants
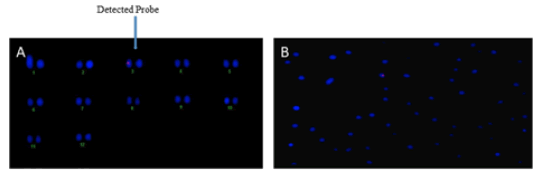
In Fluorescence In Situ Hybridization (FISH) assay where only one fluorescent signal was detected, it indicates the successful genetic integration of the MnChi10 gene into the genome of tomato. This result is pivotal for confirming the presence of MnChi10 gene in tomato chromosome, which can have significant implications for enhancing wilt resistance in tomato crops (Fig. 4). Among 180 transformed regenerated plants, only 50 plants were PCR positive. Among 50 plants 5 plants were showed stable integration of chitinase gene (Fig. 5).
Figure 4: (A) Chitinase gene integration with tomato chromosome, (B) Hybridization of probe with tomato chromosome.
Figure 5: Molecular analysis of transformed tomato plant. PCR of positive tomato plants showed chitinase gene integrated in tomato genome.
Fungal Bioessay
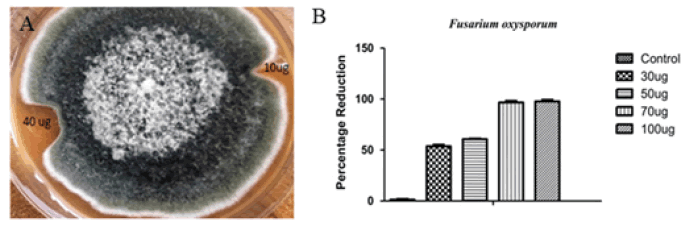
Purified and recombinant chitinase protein was used in this experiment. Fungal inhibition experiment showed that the recombinant chitinase protein of mulberry was quite effective in controlling the said fungi. Fig. 6 showed the significant retardation of growth of inoculated fungi, Fusarium oxysporum at 40 μg protein concentration and it was evident from the experiment that fungal inhibition was effective as compared to 10 μg protein concentration. In the in-vitro antifungal essay, %age reduction of hyphal growth of Fusarium oxysporum was 51.8% at protein concentration of 30 μg, 60% at 50 μg, 95% at 70 μg, 97.5% at 100 μg protein concentration. 70 μg and above protein concentrations did not allow the fungal growth across the well (Fig. 6).
Figure 6: (A) Effect of recombinant chitinase protein on growth of Fusarium oxysporum. Zone of inhibition produced by purified recombinant chitinase protein applied along the growing border of Fusarium oxysporum. (B) Percentage reduction in the hyphal growth of Fusarium oxysporum exhibited by the purified recombinant chitinase protein. The data was obtained in in-vitro quantitative antifungal assays. Control sample didn’t exhibit any reduction. The results showed that there was significant difference among control and different concentrations in standard value. (P<0.01; n=3).
Chitinase gene expression studies (RT PCR)
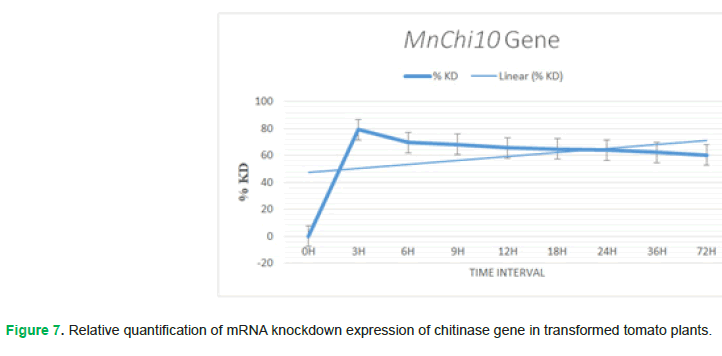
Real-time PCR is performed and results showed that the expression of chitinase gene is detected at first after infection by the pathogen than it increases within the passage of time. Increase in %KD value will decrease the gene expression, as %KD value is inversely proportional to gene expression. Initially at time interval 0 h, the Fusarium oxysporum fungi attacked tomato plant for disease infection and chitinase gene exhibits its activity but with the passage of time, in time interval 3 h, 6 h, 9 h, 12 h, 18 h, 24 h, 36 h and 72 h chitinase gene expression increases accordingly (Fig. 7).
Figure 7: Relative quantification of mRNA knockdown expression of chitinase gene in transformed tomato plants.
Purified chitinase gene structure
Primary structure:
MNMIMIHKMASSKRSCVRVLCLITLTILSCLAWPGMGTGTRSSSSPTARLISEQLFNTFFLHKDDSACPANTFYTYDAFITATKCFPRFASTGSLSTRKREIAAFLAQISHETTGGWPTAPDGPYSWGLCFKEEISPQSDYCDSSNTQWPCSPGKSYKGRGPIQLSWNYNYGPAGEALHFDGLGSPEVVANDSVVAFKTAIWFWMTPRRPKPSCHQVMVGEYVPTRDDVAANRTAGYGLVTNIINGGLECGIPNDARVNDRIGYFRRYAGLLNVDTGPNLDCENQKPF
Secondary structure: PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/&uuid=e55400e2-5afb-11ef-b81e-00163e100466) was used for protein secondary structure prediction. Following are its results (Fig. 8 and Fig. 9):
Figure 8: Chitinase gene secondary structure.
Figure 9: Extracellular or periplasmic regions, signal peptide in targeting the protein and membrane interaction in protein.
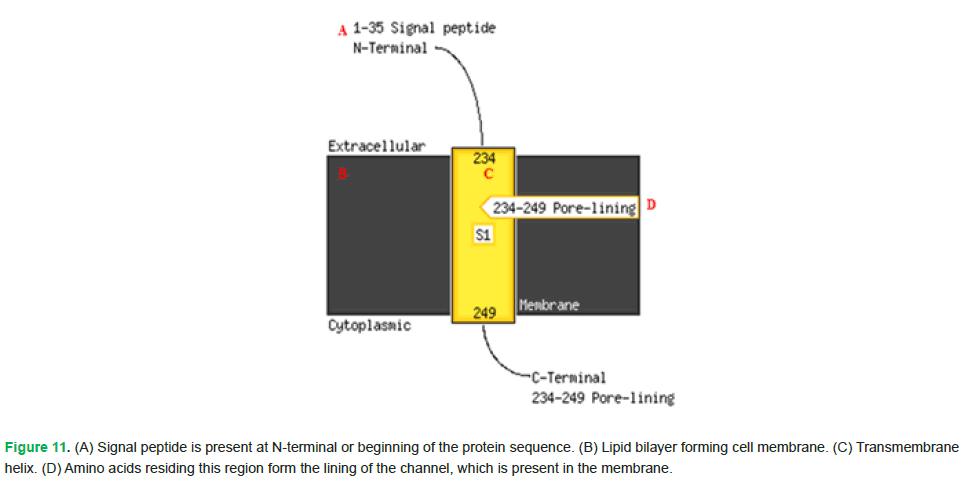
The amino acids highlighted in yellow indicate extracellular or periplasmic regions. The ones in pink denote the involvement of signal peptide in targeting the protein. Moreover, amino acids spanning from positions 234 to 249 are involved in membrane interaction (Fig. 10 and Fig. 11).
Figure 10: (A) Blue blocks represent the confidence score for each residue in the protein’s secondary structure. The higher the peaks, the higher the confidence score is, indicating a greater likelihood of the aforementioned residues being present at the desired positions. (B) Pink colored blocks represent the alpha helices. (C) It represents the fate of each residue, whether it will form alpha helix, beta strand or coil. (D) This row represents the amino acid sequence of protein. (E) Yellow colored blocks represent the beta strands.
Figure 11: (A) Signal peptide is present at N-terminal or beginning of the protein sequence. (B) Lipid bilayer forming cell membrane. (C) Transmembrane helix. (D) Amino acids residing this region form the lining of the channel, which is present in the membrane.
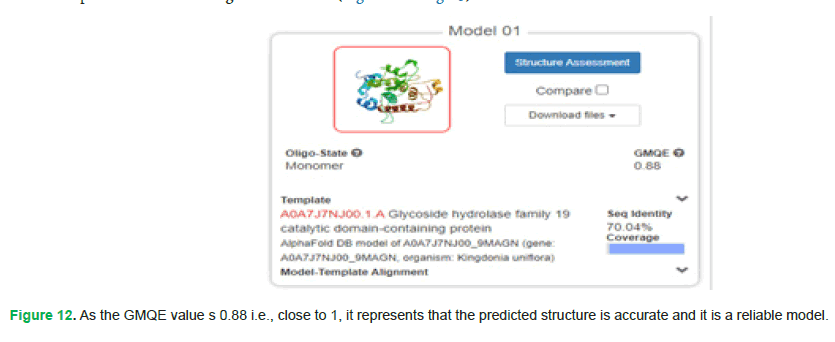
Tertiary structure: SWISS-MODEL (https://swissmodel.expasy.org/interactive/sFB2dy/models) was used for protein tertiary structure prediction. Following are its results (Fig. 12 and Fig. 13):
Figure 12: As the GMQE value s 0.88 i.e., close to 1, it represents that the predicted structure is accurate and it is a reliable model.
Figure 13: (A and B) Tertiary structures of protein.
trRosetta
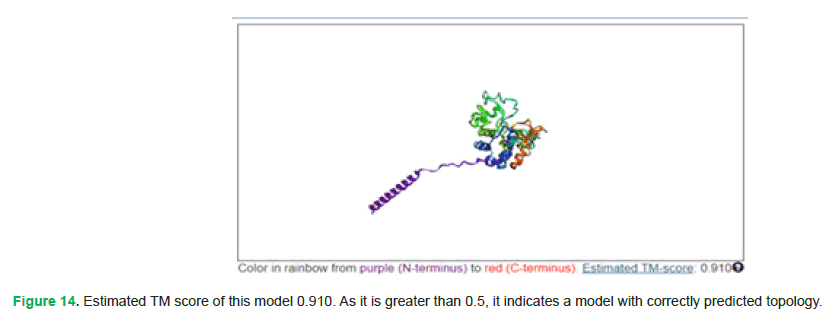
To validate the prediction of protein’s tertiary structure, trRosetta was also used (https://yanglab.qd.sdu.edu.cn/trRosetta/output/TR153286/). Following are its results (Fig. 14):
Figure 14: Estimated TM score of this model 0.910. As it is greater than 0.5, it indicates a model with correctly predicted topology.
TM score greater than 0.5 shows that the overall folding of protein is close to its native structure and has a correct topology.
Discussion
Plant chitinases are safe & biodegradable biocontrol agent that can be used in place of common fungicides (Kumar and Vivekanand, et al. 2018). Chitinases cleaves β-1,4-glucosidic bonds of chitin, which is main component in cell wall of fungi (Kumar, et al. 2018). Plants do not have the ability to produce Chitin by natural process that’s why plant chitinases has been identified to discharge elicitors for plant defense by hydrolyzing the pathogen cell wall (Hibbett, et al. 2011). Chitinase gene is effectively transformed into other plants to enhance resistance against various fungal pathogens and insects (Fisher, et al. 2012). It has been reported that various chitinases from many plants and fungi were stable to different ranges of pH and temperature (Bussink, et al. 2007). Our study aimed to evaluate the expression of chitinase gene in tomato plants and overall efficiency of MnChi10 gene to control Fusarium wilt disease. Overall functionality of the chitinase gene in transgenic product was analyzed through transfer of desired chitinase gene in tomato plant. Real time PCR was performed with transgenic plants to check chitinase gene level at various intervals of time. It showed that at time interval 0 h the chitinase gene expression in transgenic plants is detected after infection, from time interval 3 h to 72 h, the expression of chitinase gene is increasing continuously. It depicted that as chitinase gene level is increasing, it helps the plant to cope the diseased fungi. Chromosomal studies by Fluorescent In Situ Hybridization (FISH) showed that the transformation of chitinase gene in tomato plant. Chitinase gene is successfully incorporated in the genome of tomato plant. The fluorescent probe is shown on chromosome pair 3 of the total chromosomes. In Fluorescence In Situ Hybridization (FISH) assay where only one fluorescent signal was detected, it indicates the successful genetic integration of the MnChi10 gene sinto the genome of tomato. This result is pivotal for confirming the presence of MnChi10 gene in tomato chromosomes, which can have significant implications for enhancing disease resistance in tomato crops. Chitinases genes are involved in plant safety against damaging organisms/plant pathogens (Downing and Thomson, 2000). Most of the transformed plants showed mild symptoms upon infection or didn`t showed symptoms which showed successful integration of chitinase gene in tomato genome (Kabir, et al. 2016).chitinases are revolutionary in agriculture sector because they alter the use of chemicals on the plants (Kumar and Vivekanand, et al. 2018, Dean, et al. 2012). Our results documented the transgenic plants with chitinase gene MnChi10 recorded enhanced resistance against Fusarium wilts.
Conclusion
In conclusion, plant chitinases can be widely used to control fungal diseases and increase the resistance of plants. The tomato plant showed greater flexibility for incorporation the MnChi10 gene from the. MnChi10 gene showed promising results to build resistance in tomato against wilt fungi. There is a strong need to commercialize the transgenic tomato to support farmers’ livelihoods and increase national income to boost the economy.
Acknowledgement
The Author is highly obliged to respected faculty and supporting staff of Faculty of Agricultural Sciences and Center of Excel - lence in Molecular Biology, University of the Punjab Lahore Pakistan for support and necessary equipment needed.
Funding
All crop material, animals, chemicals, and materials provided by Faculty of Agriculture Sciences.
Consent to Publish
All the authors declared they have no objection to publish manuscript.
References
- Mwangi RW, Mustafa M, Charles K, Wagara IW, Kappel N. (2023). Selected emerging and reemerging plant pathogens affecting the food basket: A threat to food security. J Agr Food Res. 14:100827.
- Doehlemann G, Ökmen B, Zhu W, Sharon A. (2017). Plant pathogenic fungi. The Fungal Kingdom. 701-726.
- Fauteux F, Rémus-Borel W, Menzies JG, Bélanger RR. (2005). Silicon and plant disease resistance against pathogenic fungi. FEMS Microbiol Let. 249:1-6.
[Crossref] [Google Scholar] [PubMed]
- Thambugala KM, Daranagama DA, Phillips AJ, Kannangara SD, Promputtha I. (2020). Fungi vs. fungi in biocontrol: An overview of fungal antagonists applied against fungal plant pathogens. Front Cell Infect Microbiol. 10:604923.
[Crossref] [Google Scholar] [PubMed]
- Panno S, Davino S, Caruso AG, Bertacca S, Crnogorac A, MandiÄ A, Noris E, MatiÄ S. (2021). A review of the most common and economically important diseases that undermine the cultivation of tomato crop in the mediterranean basin. Agron. 11:2188.
- Brahimi M, Boukhalfa K, Moussaoui A. (2017). Deep learning for tomato diseases: Classification and symptoms visualization. Applied Artificial Intelligence. 31:299-315.
- Staniaszek M, Szczechura W, Marczewski W. (2014). Identification of a new molecular marker C2-25 linked to the Fusarium oxysporum f. sp. radicis-lycopersici resistance Frlgene in tomato. Czech J Genet Plant Breed. 50:285-287
- Ullah N, Ali AS, Ahmad M, Fahim M, Din N, Ahmad F. (2017). Evaluation of tomato genotypes against Tomato Mosaic Virus (ToMV) and its effect on yield contributing parameters. Pak J Bot. 49:1585-1592.
- Collins EJ, Bowyer C, Tsouza A, Chopra M. (2022). Tomatoes: An extensive review of the associated health impacts of tomatoes and factors that can affect their cultivation. Biology. 11:239.
[Crossref] [Google Scholar] [PubMed]
- Nazarov PA, Baleev DN, Ivanova MI, Sokolova LM, Karakozova MV. (2020). Infectious plant diseases: Etiology, current status, problems and prospects in plant protection. Acta Naturae. 12:46.
[Crossref] [Google Scholar] [PubMed]
- Chitwood-Brown J, Vallad GE, Lee TG, Hutton SF. (2021). Breeding for resistance to Fusarium wilt of tomato: A review. Genes. 12:1673.
[Crossref] [Google Scholar] [PubMed]
- Arie T. (2019). Fusarium diseases of cultivated plants, control, diagnosis, and molecular and genetic studies. J Pestic Sci. 44:275-81.
[Crossref] [Google Scholar] [PubMed]
- Wang CJ, Thanarut C, Sun PL, Chung WH. (2020). Colonization of human opportunistic Fusarium oxysporum (HOFo) isolates in tomato and cucumber tissues assessed by a specific molecular marker. PLoS One. 15:e0234517.
[Crossref] [Google Scholar] [PubMed]
- Jiménez Díaz RM, Jiménez-Gasco MD. (2011). Integrated management of Fusarium wilt diseases. 177-215.
- Recep K, Fikrettin S, Erkol D, Cafer E. (2009). Biological control of the potato dry rot caused by Fusarium species using PGPR strains. Biological control. 50:194-198.
- Chaerani R, Voorrips RE. (2006). Voorrips, Tomato early blight (Alternaria solani): The pathogen, genetics, and breeding for resistance. J Gen Plant Pathol. 72:335-347.
- Anderson PK, Cunningham AA, Patel NG, Morales FJ, Epstein PR, Daszak P. (2004). Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol Evol. 19:535-544.
[Crossref] [Google Scholar] [PubMed]
- Hamid R, Khan MA, Ahmad M, Ahmad MM, Abdin MZ, Musarrat J, Javed S. (2013). Chitinases: An update. J Pharm Bioallied Sci. 5:21-29.
[Crossref] [Google Scholar] [PubMed]
- Rathore AS, Gupta RD. (2015). Chitinases from bacteria to human: Properties, applications, and future perspectives. Enzyme Res. 2015:791907.
[Crossref] [Google Scholar] [PubMed]
- Wubie AJ, Hu Y, Li W, Huang J, Guo Z, Xu S, Zhou T. (2014). Factors analysis in protoplast isolation and regeneration from a chalkbrood fungus, Ascosphaera apis. Int J Agric Biol. 16:89-96.
- García PC, Rivero RM, López-Lefebre LR, Sánchez E, Ruiz JM, Romero L. (2001). Response of oxidative metabolism to the application of carbendazim plus boron in tobacco. Funct Plant Biol. 28:801-806.
- Suginta W, Robertson PA, Austin B, Fry SC, FothergillâGilmore LA. (2000). Chitinases from Vibrio: Activity screening and purification of chiA from Vibrio carchariae. J Appl Microbiol. 89:76-84.
[Crossref] [Google Scholar] [PubMed]
- Dana MD, Pintor-Toro JA, Cubero B. (2006). Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiol. 142:722-730.
[Crossref] [Google Scholar] [PubMed]
- Kumar M, Brar A, Yadav M, Chawade A, Vivekanand V, Pareek N. (2018). Chitinases-potential candidates for enhanced plant resistance towards fungal pathogens. Agric. 8:88.
- Cletus J, Balasubramanian V, Vashisht D, Sakthivel N. (2013). Transgenic expression of plant chitinases to enhance disease resistance. Biotechnol Lett. 35:1719-1732.
[Crossref] [Google Scholar] [PubMed]
- Sivaji M, Sadasivam V, Narayanasamy J, Samuel S, Fan C. (2014). Detection, characterization and evolution of internal repeats in chitinases of known 3-D structure. PLoS One. 9:e91915.
[Crossref] [Google Scholar] [PubMed]
- Cao J, Tan X. (2019). Comprehensive analysis of the chitinase family genes in tomato (Solanum lycopersicum). Plants. 8:52.
[Crossref] [Google Scholar] [PubMed]
- Collinge DB, Munk L, Cooke BM. (2008). Sustainable disease management in a European context. Eur J Plant Pathol. 121:213-409.
- Davis JM, Wu H, Cooke JE, Reed JM, Luce KS, Michler CH. (2002). Pathogen challenge, salicylic acid, and jasmonic acid regulate expression of chitinase gene homologs in pine. Mol Plant Microbe Interact. 15:380-387.
[Crossref] [Google Scholar] [PubMed]
- Di X, Takken FL, Tintor N. (2016). How phytohormones shape interactions between plants and the soil-borne fungus Fusarium oxysporum. Front Plant Sci. 7:170.
[Crossref] [Google Scholar] [PubMed]
- Gai YP, Zhao YN, Zhao HN, Yuan CZ, Yuan SS, Li S, Zhu BS, Ji XL. (2017). The latex protein MLX56 from mulberry (Morus multicaulis) protects plants against insect pests and pathogens. Front Plant Sci. 8:1475.
[Crossref] [Google Scholar] [PubMed]
- Kumar M, Vivekanand V, Pareek N. (2018). Structure, regulation, and potential applications of insect chitin-metabolizing enzymes. Trends in Insect Mol Biol Biotechnol. 2018:295-316.
- Hibbett DS, Ohman A, Glotzer D, Nuhn M, Kirk P, Nilsson RH. (2011). Progress in molecular and morphological taxon discovery in Fungi and options for formal classification of environmental sequences. Fungal Biol Rev. 25:38-47.
- Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature. 484:186-194.
[Crossref] [Google Scholar] [PubMed]
- Bussink AP, Speijer D, Aerts JM, Boot RG. (2007). Evolution of mammalian chitinase (-like) members of family 18 glycosyl hydrolases. Genetics. 177:959-70.
[Crossref] [Google Scholar] [PubMed]
- Downing KJ, Thomson JA. (2000). Introduction of the Serratia marcescens chiA gene into an endophytic Pseudomonas fluorescens for the biocontrol of phytopathogenic fungi. Can J Microbiol. 46:363-9.
[Crossref] [Google Scholar] [PubMed]
- Kabir SR, Rahman MM, Tasnim S, Karim MR, Khatun N, Hasan I, Amin R, Islam SS, Nurujjaman M, Kabir AH, Sana NK. (2016). Purification and characterization of a novel chitinase from Trichosanthes dioica seed with antifungal activity. Int J Biol Macromol. 84:62-8.
[Crossref] [Google Scholar] [PubMed]
- Dean R, Van Kan JA, Pretorius ZA, HammondâKosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, Foster GD. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 13:414-30.
[Crossref] [Google Scholar] [PubMed]
Copyright: Mubarak Ali Anjum, Department of Plant Pathology, Faculty of Agricultural Sciences, University of the Punjab, Lahore, Pakistan, Email: mubarak.ali.anjum1103@gmail.com This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.